Explanation of parameters and their effects#
Note
The example below pertains to the segmentation of actin filaments. Your options will vary depending on the classical segmentation workflow chosen within the Allen Cell napari plugin.
Pre-processing: Intensity normalization#
Intended for scaling the intensity of the image between 0-1. The parameters are scaling_param 1 and scaling_param 2. These set the range from the mean image intensity to normalised.
The mean image intensity is calculated for the image; then scaling_param 1 and 2 are subtracted or added to the mean, setting a range that will be scaled from 0-1.
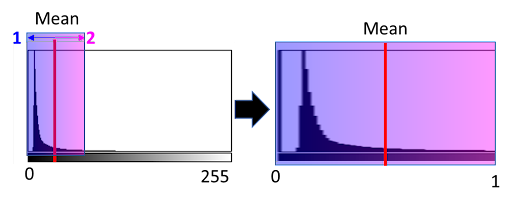
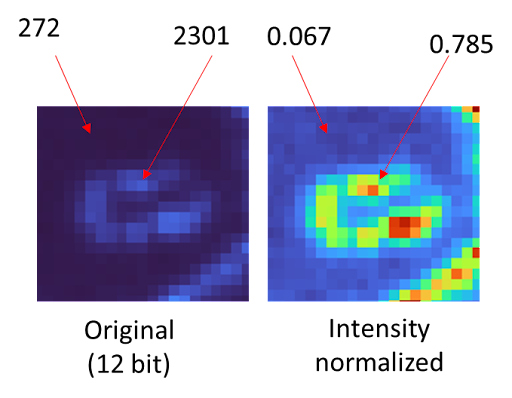
Parameters here will decide how much of the histogram you want to exclude as being too bright (i.e. too many dead cells), etc.
Hint
Values dependent on the image mean value and intensity skew in the image itself.
Pre-processing: Edge preserving smoothing#
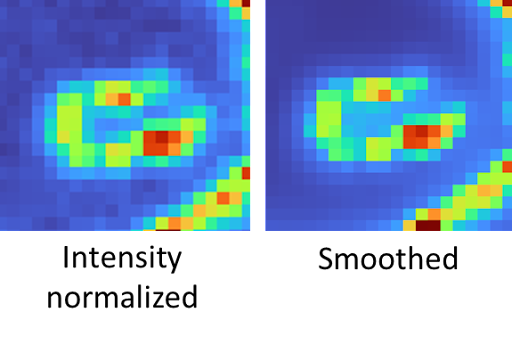
An image is smoothed to reduce noise. No parameters are required.
Core segmentation: Sigma#
Sigma allows you to set the expected thickness of filaments. Smaller values lend themselves to thinner filaments; while larger values are best used with thicker filaments.
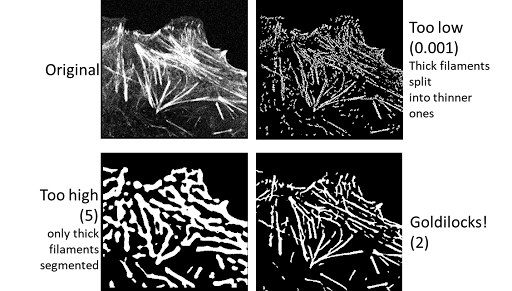
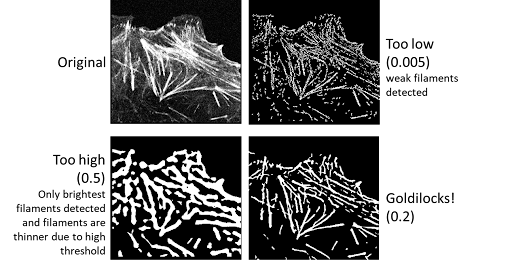
Core segmentation: Cutoff#
Cutoff allows you to set the threshold for the image.
Post-processing#
Size filtering can be used to remove spurious segmentations. You can link segmentations in 2D or 3D using the connecting pixels controls.
Note
If you find in using this plugin that you have quite disparate structures, the same workflow can be run twice to segment different populations within your data.
