AreTomoLive Demo using data from the CryoET Data Portal¶
This guide introduces the AreTomoLive [1] pipeline to perform cryoET data preprocessing, spanning motion correction of the acquired tilt-movies to contrast enhancement of the reconstructed tomograms. The AreTomoLive pipeline consists of two GPU-accelerated packages, AreTomo3 and DenoisET. AreTomo3 provides a fully automated, multi-GPU-accelerated workflow to reconstruct tomograms from raw tilt-series with quality metrics reported. DenoisET implements the machine learning algorithm Noise2Noise and is designed to run in parallel with AreTomo3 to perform contrast enhancement. This demo provides instructions for running both packages on a dataset of purified synaptosomes, which is available on the CryoET Data Portal.
Background Information¶
Dataset¶
The dataset of purified synaptosomes used in this demo is available on the CryoET Data Portal, with some additional details below:
The sample of purified synaptosomes was generated from rat hippocampi.
The full dataset includes 76 runs. Each run contains the raw movies, the motion-corrected tilt-series, two types of tomograms, and membrane segmentations. Only the raw movies are needed to test AreTomoLive.
The tomograms on the CryoET Data Portal were also reconstructed by AreTomo3 and denoised by DenoisET so can be compared with the tomograms from this demo.
The options to download this dataset are:
Software¶
Both software packages used in this tutorial are publicly available on GitHub:
AreTomo3: https://github.com/czimaginginstitute/AreTomo3
The latest version on GitHub and in this demo is 2.1.3. You can download a pre-compiled executable from GitHub.
DenoisET: https://github.com/apeck12/denoiset
The latest version on GitHub is 0.1.0. Dependencies are specified in the
pyproject.tomlfile.
Hardware Requirements¶
A Linux workstation or Linux HPC equipped with at least 1 NVIDIA GPU card
Minimum CUDA version: 12.0.0
Minimum CPU RAM: 64 GB x number of GPUs for AreTomo3, 96 GB on 1 GPU for DenoisET
For the data in this tutorial, minimum disk storage: ∼20 GB for 10 tilt series or ∼152 GB for the full dataset (76 tilt series)
Installation¶
For AreTomo3, a pre-compiled executable is provided with this tutorial. Alternatively, the GitHub link provides instructions for how to compile the code from scratch.
For DenoisET, the following commands install the code in a new conda environment on a GPU-available machine:
git clone https://github.com/apeck12/denoiset.git
cd denoiset
conda create \--name denoiset python=3.11.4
conda activate denoiset
pip install .
Demo Instructions¶
Step 0: Download the dataset¶
Download the tilt-series frames in .eer format, gain reference, and mdoc files to a flat directory structure. Each
tilt series is about 2 GB.
If only running DenoisET in live inference mode with a pre-trained model, download 10 tilt-series.
If training a new denoising model from scratch, we recommend downloading the full dataset, which contains 76 tilt-series.
Example bash code for using AWS-CLI to download the full dataset to $DESTINATION_PATH. Downloading these 76
tilt-series with ∼1GB/s download speed takes approximately 30 minutes.
#!/bin/bash
DESTINATION\_PATH="/processing/basepath/raw\_data"
for dir in $(aws s3 --no-sign-request ls s3://cryoet-data-portal-public/10447/ --recursive | \
grep '/Frames/' | awk '{print $4}' | cut \-d'/' -f1-3 | sort -u ); do
# Specify the flat destination directory
local_dir="$DESTINATION_PATH"
# Create the destination directory if it does not exist
mkdir -p "$local_dir"
# Sync files from S3 to local directory in flat structure
aws s3 sync "s3://cryoet-data-portal-public/$dir" "$local_dir" --no-sign-request \
--exact-timestamps
done
Also download the gain reference to DESTINATION_GAIN_PATH.
DESTINATION_GAIN_PATH="/processing/basepath/gain\_ref"
aws s3 sync "s3://cryoet-data-portal-public/10447/24sep26a_Position_1/Gains" \
"$DESTINATION_GAIN_PATH" --no-sign-request --exact-timestamps
Step 1: Run AreTomo3¶
To check the AreTomo3 executable and see the help menu, run:
/path/to/aretomo3_executable --help
Starting from raw tilt-series frames in a flat directory structure ($IN_PREFIX), the following AreTomo3 command
applies both local and global corrections for beam-induced motion (BIM) in 2D and 3D. Tomograms are then reconstructed
by weighted backprojection, with a local correction for the contrast transfer function (CTF) applied:
IN_PREFIX="/processing/basepath/raw_data/24sep26a_Position_"
OUT_DIR="/processing/basepath/aretomo3_output"
GAIN_PATH="/processing/basepath/gain_ref/20240924_101412_EER_GainReference.gain"
/path/to/AreTomo3_2.1.3_03-19-2025 -InPrefix $IN_PREFIX -InSuffix .mdoc -OutDir $OUT_DIR \
-Gain $GAIN_PATH -PixSize 1.54 -kV 300 -Cs 2.7 -SplitSum 1 -McPatch 5 5 -McBin 1 -Group 2 4 \
-AtPatch 4 4 -AtBin 3.25 -OutImod 1 -Wbp 1 -CorrCTF 1 15 -FlipVol 1 -Gpu 0,1,2,3,4,5,6,7 \
-Serial 1000 2>/dev/null
The instruction manual on GitHub contains more details about the command-line arguments, but below we provide a few notes about the most frequently adjusted arguments:
If denoising using a pretrained model, modify the above to:
-SplitSum 0to prevent writing out even (*EVN Vol.mrc) and odd (*ODD Vol.mrc) volumes.The pixel size of the reconstruction is the product of
PixSize,McBin, andAtBin, which correspond to the pixel size of the raw frames, the bin factor to generate the motion-corrected tilt-series, and the bin factor used during reconstruction. The above command reconstructs tomograms with a 5 Å pixel size.
On a Linux server equipped with 8 NVIDIA RTX A6000 GPUs, AreTomo3 finished processing all 76 tilt-series in approximately 3 hours.
Step 2: Run DenoisET¶
Once in the denoiset conda environment, the following command checks the installation and provides a list of command-line arguments:
denoise3d --help
The following command runs denoising live in parallel with AreTomo3:
input="/processing/basepath/aretomo3_output"
output="/processing/basepath/denoiset_output_live_train"
denoise3d \
--input ${input} \
--output ${output} \
--metrics_file ${input}/TiltSeries_Metrics.csv \
--tilt_axis 1.0 \
--global_shift 400 \
--ctf_res 10 \
--ctf_score 0.2 \
--bad_patch_low 0.01 \
--bad_patch_all 0.05 \
--min_selected 25 \
--live \
--t_exit 7200
Since the default thresholds for the metrics-related arguments were set based on our experience with lamella data, these
thresholds are adjusted above to be more selective for the synaptosome dataset. This command can be run immediately
after AreTomo3 processing starts. On a single NVIDIA RTX A40 GPU, training and inference finished in 7 hours when we ran
this command concurrently with AreTomo3, which includes the time spent monitoring for sufficient high-quality tomograms
to use for training. However, training time is stochastic and will vary between runs. If the output tomograms appear to
be insufficiently denoised, we recommend increasing the min_selected or n_extract (default: 250) parameters.
Alternatively, if a suitable pretrained model is available, inference can run live in parallel with AreTomo3:
input="/processing/basepath/aretomo3_output"
output_inference="/processing/basepath/denoiset_output"
model="/path/to/denoiset/models/synaptosome.pth"
predict3d --input ${input} --output ${output_inference} --model ${model} --live --t_exit 7200
On a single NVIDIA RTX A40 GPU, inference for all 76 tilt-series using this pretrained model finished in ap proximately 3 hours when run concurrently with AreTomo3 and 2 hours when run separately after AreTomo3 had already finished.
Expected Output¶
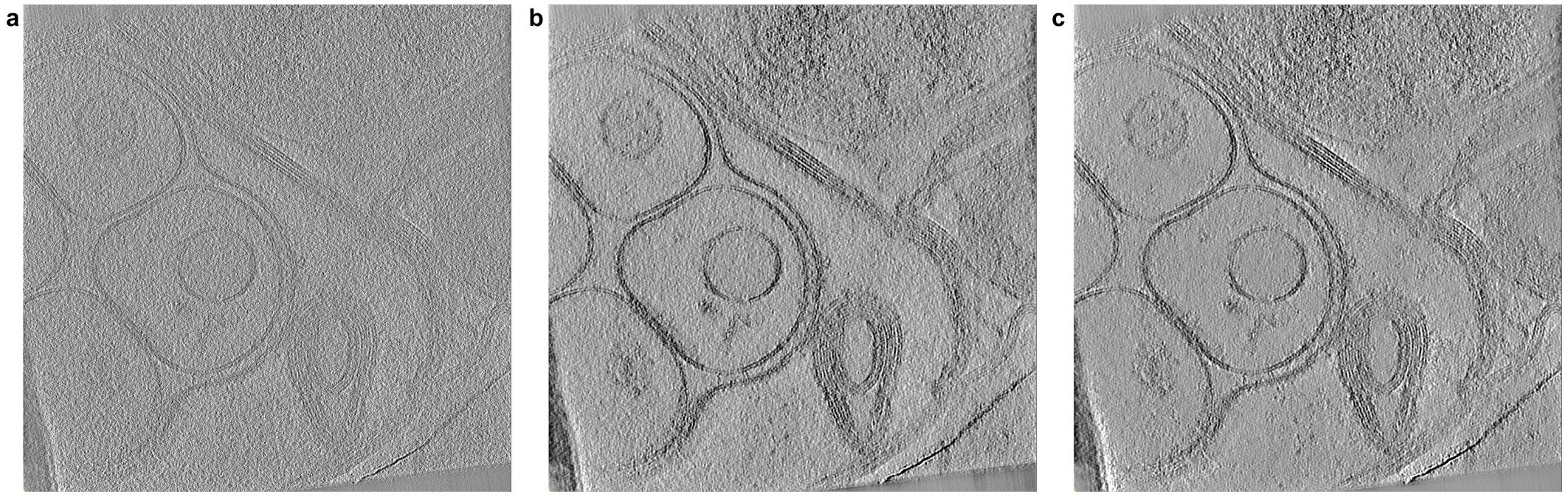
The same slice is visualized through a. the CTF-deconvolved tomogram reconstructed by AreTomo3 and this tomogram after denoising with b. the provided pre-trained model or c. the new model trained from scratch.¶

The same slice is visualized through a. the CTF-deconvolved tomogram reconstructed by AreTomo3 and this tomogram after denoising with b. the provided pre-trained model or c. the new model trained from scratch.¶
AreTomo3 output files¶
The following files will be found in the main output folder:
General files
AreTomo3_Session.json: record of the run parametersTiltSeries_Metrics.csv: record of the tilt-series quality metricsMdocDone.txt: list of the processed tilt-seriesTiltSeries_TimeStamp.csv: list of processing timestamps
Per tilt-series (run) files
{run}.aln: record of global and local alignments{run}.mrc: 2D motion-corrected tilt-series (tomographically unaligned){run}CTF.mrc,{run}CTF.txt: CTF fits and parameters{run}_Vol.mrc: reconstructed volume
Subfolders
{run}_IMOD: contains IMOD-style CTF and alignment files{run}_Log: stores log files for the processed tilt-series
DenoisET output files¶
The following files will be found in the main output folder:
denoise3d.jsonorpredict3d.json: record of the run parameters{run}_Vol.mrc: denoised tomogramtraining: if training from scratch, directory containing the following files:
epoch{n}.pth: model weights from the nth epoch{run}_epoch{n}.mrc: representative denoised tomogram after the nth epochtraininglist.txt: list of tomograms selected for trainingtraining_stats.csv: per epoch statistics like training losstraininglist.png: figure comparing distribution of quality metrics for the selected and all tomograms
Contact¶
For questions or comments, please contact shawn.zheng@czii.org or ariana.peck@czii.org.
References¶
[1] Ariana Peck, Yue Yu, Mohammadreza Paraan, Dari Kimanius, Utz Heinrich Ermel, Joshua Hutchings, Daniel Serwas, Hannah Siems, Norbert S Hill, Mallak Ali, et al. Aretomolive: Automated reconstruction of comprehensively-corrected and denoised cryo-electron tomograms in real-time and at high throughput. bioRxiv, pages 2025–03, 2025.