Data Organization¶
The Data Portal is organized in a hierarchical structure. We welcome contributions to the Portal. Use this form to express interest in adding your data to the Portal.
Overview¶
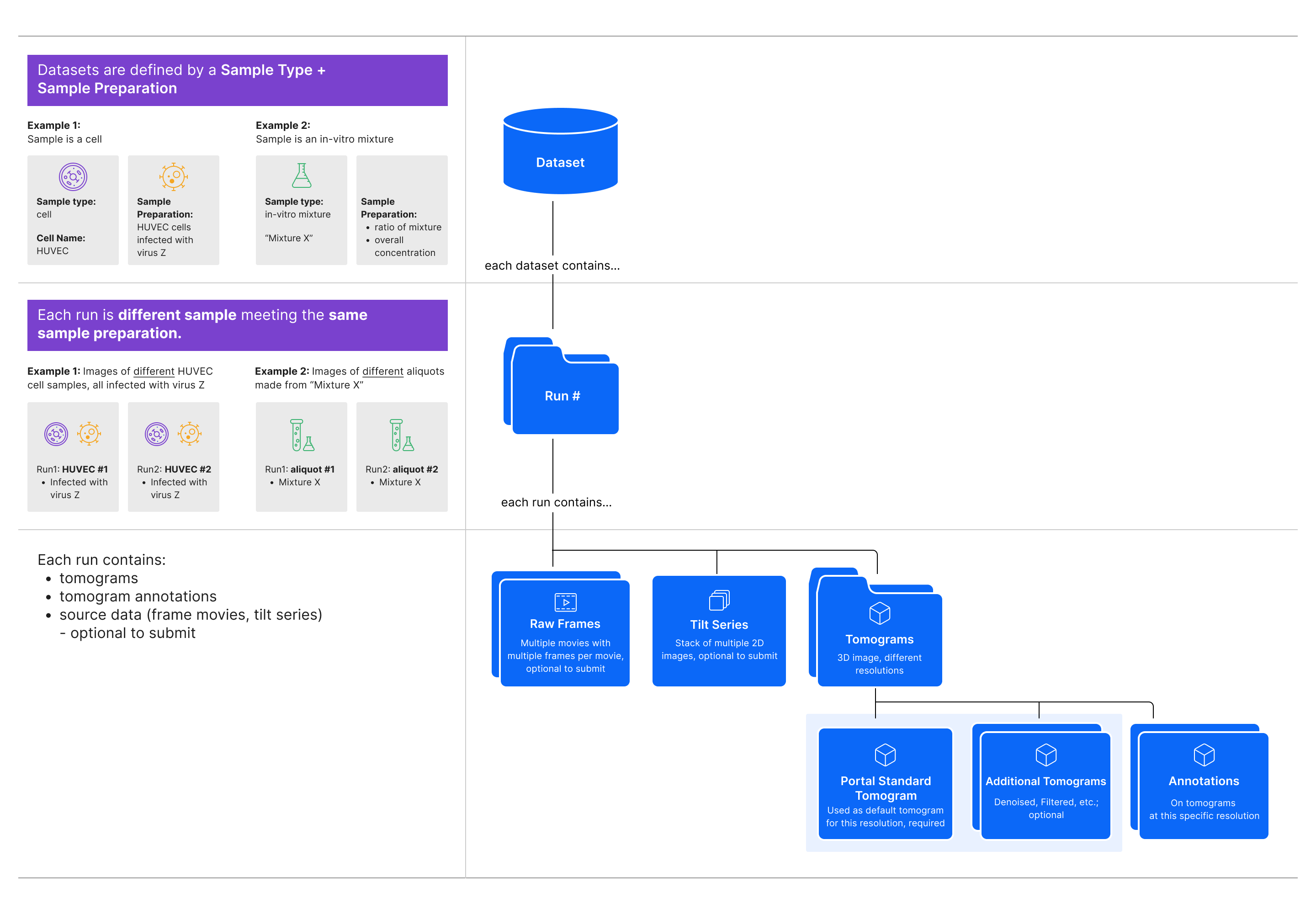
CryoET Data Portal Data Hierarchy¶
As shown in the diagram above, the CryoET Data Portal has 3 levels in the data hierarchy:
Dataset is a set of image files for tilt series, reconstructed tomograms, and cellular and/or subcellular annotation files. Every dataset contains only one sample type prepared with the same conditions. The dataset title, such as “S. pombe cryo-FIB lamellae acquired with defocus-only,” summarizes these conditions. Samples can be a cell, tissue or organism; intact organelle; in-vitro mixture of macromolecules or their complex; or in-silico synthetic data, where the experimental conditions are kept constant. Datasets typically contain multiple runs (see below). Downloading a dataset downloads all files, including all available tilt series, tomograms, and annotations.
Run is one experiment, or replicate, within a dataset, where all runs in a dataset have the same sample conditions. Every run contains a collection of all tomography data and annotations related to imaging one physical location in a sample. It typically contains one tilt series and all associated data (e.g. movie frames, tilt series image stack, tomograms, annotations, and metadata), but in some cases, it may be a set of tilt series that form a mosaic. Runs contain at least one annotation for every tomogram. When downloading a run from a Portal page, you may choose to download the tomogram or all available annotations. To download all data associated with a run (i.e. all available movie frames, tilt series image stack, tomograms, annotations, and associated metadata), use the API.
Annotation is a point or segmentation indicating the location of a macromolecular complex in the tomogram. On a run overview page, you may choose to download individual annotations.
All data is added to the Portal through Depositions, which is described below, and a subset of depositions are displayed in the depositions tab on the Portal.
For more detailed explanations refer to the sections below.
Depositions¶
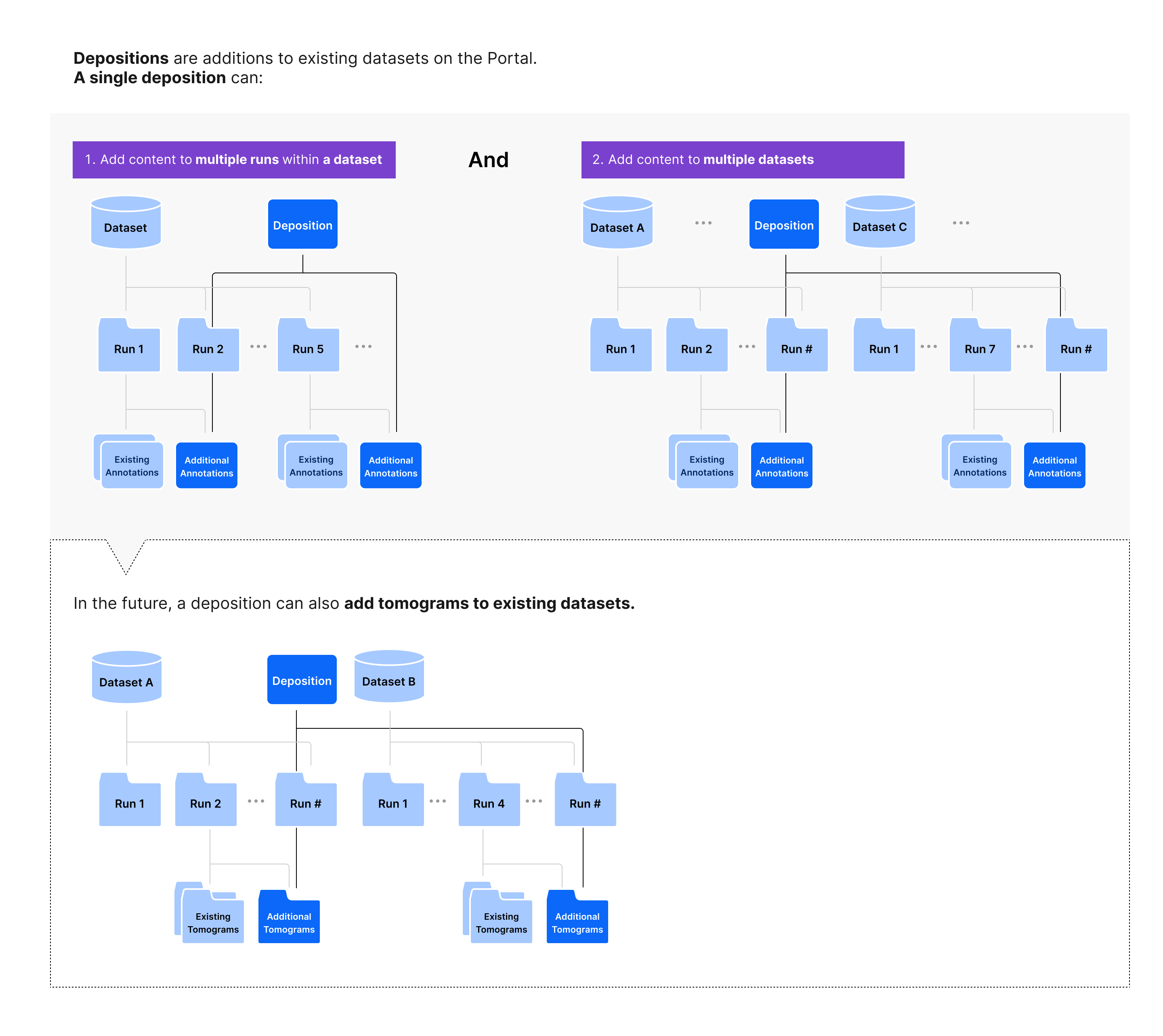
Types of depositions¶
Depositions are collections of data submitted together. All data being submitted together will be tagged with the same deposition ID, starting with CZCDP (e.g. CZCDP-12345; note that only the numeric part is supported in the API). On the Portal, we will surface depositions that contain annotations submitted together. In the future, depositions surfaced on the Portal may include tomograms added to existing datasets or sets of datasets contributed together.
Datasets¶
Datasets are contributed sets of image files associated with imaging one sample type with the same sample preparation methods. Datasets contain runs, where each run corresponds to imaging one physical location in the prepared samples. Dataset IDs start with DS (e.g. DS-10000; note that only the numeric part is supported in the API).
The Browse Datasets page shows a table of all datasets on the Portal. These datasets are not currently ordered. Instead, the left side filter panel provides options for filtering the table according to files included in the datasets, such as ground truth annotation files; the author or ID of the dataset; organism in the sample; hardware; metadata for the tilt series or reconstructed tomograms. In addition, the search bar filters based on keywords or phrases contained in the dataset titles. The dataset entries in the table have descriptive names, such as “S. pombe cryo-FIB lamellae acquired with defocus-only,” which aim to summarize the experiment as well as a Dataset ID assigned by the Portal, the organism name, number of runs in the dataset, and list of annotated objects, such as membrane. Datasets on the Portal may be found in other image databases. On the Browse Datasets page, the datasets table shows the EMPIAR ID for datasets that are also found on the Electron Microscopy Public Image Archive.
On a given Dataset Overview page, the View All Info panel contains metadata for the dataset. These metadata are defined in the tables below including their mapping to attributes in the Portal API:
Dataset Metadata
Portal Metadata |
API Expression |
Definition |
|---|---|---|
Deposition Date |
Dataset.deposition_date |
Date when a dataset is initially received by the Data Portal. |
Grant ID |
DatasetFunding.grant_id |
Grant identifier provided by the funding agency. |
Funding Agency |
DatasetFunding.funding_agency_name |
Name of the funding agency. |
Related Databases |
Dataset.related_database_entries |
The dataset identifier for other databases, e.g. EMPIAR, that contain this dataset. |
Sample and Experiment Conditions
Portal Metadata |
API Expression |
Definition |
|---|---|---|
Sample Type |
Dataset.sample_type |
Type of sample: cell, tissue, organism, intact organelle, in-vitro mixture, in-silico synthetic data, other. |
Organism Name |
Dataset.organism_name |
Name of the organism from which the biological sample is derived from, e.g. homo sapiens. |
Tissue Name |
Dataset.tissue_name |
Name of the tissue from which a biological sample used in a CryoET study is derived from. |
Cell Name |
Dataset.cell_name |
Name of the cell from which a biological sample used in a CryoET study is derived from, e.g. sperm. |
Cell Line or Strain Name |
Dataset.cell_strain_name |
Cell line or strain for the sample e.g. C57BI |
Cellular Component |
Dataset.cell_component_name |
Name of the cellular component, e.g. sperm flagellum |
Sample Preparation |
Dataset.sample_preparation |
Description of how the sample was prepared. |
Grid Preparation |
Dataset.grid_preparation |
Description of how the CryoET grid was prepared. |
Other Setup |
Dataset.other_setup |
Description of other setup not covered by sample preparation or grid preparation that may make this dataset unique in the same publication. |
Tilt Series
Portal Metadata |
API Expression |
Definition |
|---|---|---|
Acceleration Voltage |
TiltSeries.acceleration_voltage |
Electron Microscope Accelerator voltage in volts. |
Spherical Aberration Constant |
TiltSeries.spherical_aberration_constant |
Spherical Aberration Constant of the objective lens in millimeters. |
Microscope Manufacturer |
TiltSeries.microscope_manufacturer |
Name of the microscope manufacturer. |
Microscope Model |
TiltSeries.microscope_model |
Microscope model name. |
Energy Filter |
TiltSeries.microscope_energy_filter |
Energy filter setup used. |
Phase Plate |
TiltSeries.microscope_phase_plate |
Phase plate configuration. |
Image Corrector |
TiltSeries.microscope_image_corrector |
Image corrector setup. |
Additional microscope optical setup |
TiltSeries.microscope_additional_info |
Other microscope optical setup information, in addition to energy filter, phase plate and image corrector. |
Camera Manufacturer |
TiltSeries.camera_manufacturer |
Name of the camera manufacturer. |
Camera Model |
TiltSeries.camera_model |
Camera model name. |
Dataset Overview Page¶
The table on a Dataset Overview page contains an overview of the runs in the dataset. Each run has a name defined by the dataset authors as well as a Run ID, which is assigned by the Portal and is subject to change in the rare case where the run data needs to be re-ingested to the Portal. In addition to the run name, the tilt series quality and list of annotated objects is detailed for each run. Each entry has a View Tomogram button which will display the tomogram using an instance of Neuroglancer in the browser. The tomogram is visualized along with annotations that are manually chosen to display as many annotations as possible without overlap or occlusion.
Tilt Series Quality¶
The tilt series quality score is assigned by the dataset authors to communicate their quality estimate to users. This score is only applicable for comparing tilt series within a dataset on a relative subjective scale. Score ranges 1-5, with 5 being best. Below is an example scale based mainly on alignability and usefulness for the intended analysis.
Rating |
Quality |
Description |
|---|---|---|
5 |
Excellent |
Full Tilt Series/Reconstructions could be used in publication ready figures. |
4 |
Good |
Full Tilt Series/Reconstructions are useful for analysis (subtomogram averaging, segmentation). |
3 |
Medium |
Minor parts of the tilt series (projection images) need to be or have been discarded prior to reconstruction and analysis. |
2 |
Marginal |
Major parts of the tilt series (projection images) need to be or have been discarded prior to reconstruction and analysis. Useful for analysis only after heavy manual intervention. |
1 |
Low |
Not useful for analysis with current tools (not alignable), useful as a test case for problematic data only. |
Dataset Download Options¶
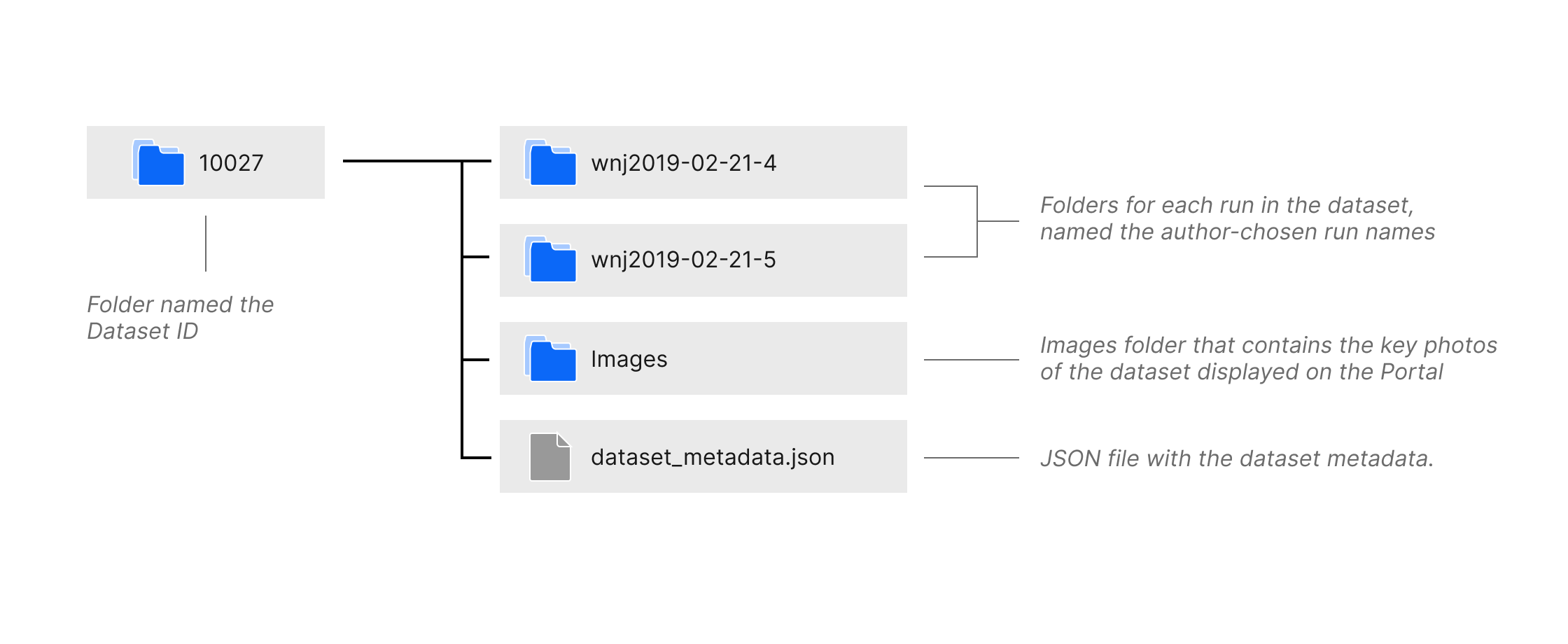
File Structure of a downloaded dataset¶
The Download Dataset button opens a dialog with instructions for downloading the dataset using Amazon Web Services Command Line Interface or the Portal API. Datasets are downloaded as folders named the Dataset ID. As shown in the diagram above, the folder contains subfolders for each run named the author-chosen run name, a folder named Images which contains the key photos of the dataset displayed on the Portal, and a JSON file named dataset_metadata.json containing the dataset metadata. The run folders contain subfolders named Tomogram and TiltSeries, containing the tomogram and tilt series image files, and a JSON file named run_metadata.json containing the run metadata. More details on the run folder file structure is found in the documentation below.
The metadata schema of any JSON file stored with the data on the data portal’s S3 bucket is described in LinkML and can be found here.
Runs¶
A tomography run is a collection of all data and annotations related to one physical location in a sample and is associated with a dataset that typically contains many other runs. On the Data Portal pages, runs are directly linked to their tomograms. For a single run, multiple tomograms of different spacings can be available. Run IDs start with RN (e.g. RN-427; note that only the numeric part is supported in the API).
An overview of all runs in a dataset is presented in the Dataset Overview page. Each run has its own Run Overview Page, where the View All Info panel contains metadata for the run. These metadata are defined in the tables below including their mapping to attributes in the Portal API:
Tilt Series
Portal Metadata |
API Expression |
Definition |
|---|---|---|
Microscope Manufacturer |
TiltSeries.microscope_manufacturer |
Name of the microscope manufacturer. |
Microscope Model |
TiltSeries.microscope_model |
Microscope model name. |
Phase Plate |
TiltSeries.microscope_phase_plate |
Phase plate configuration. |
Image Corrector |
TiltSeries.microscope_image_corrector |
Image corrector setup. |
Additional microscope optical setup |
TiltSeries.microscope_additional_info |
Other microscope optical setup information, in addition to energy filter, phase plate and image corrector. |
Acceleration Voltage |
TiltSeries.acceleration_voltage |
Electron Microscope Accelerator voltage in volts. |
Spherical Aberration Constant |
TiltSeries.spherical_aberration_constant |
Spherical Aberration Constant of the objective lens in millimeters. |
Camera Manufacturer |
TiltSeries.camera_manufacturer |
Name of the camera manufacturer. |
Camera Model |
TiltSeries.camera_model |
Camera model name. |
Energy Filter |
TiltSeries.microscope_energy_filter |
Energy filter setup used. |
Data Acquisition Software |
TiltSeries.data_acquisition_software |
Software used to collect data. |
Pixel Spacing |
TiltSeries.pixel_spacing |
Pixel spacing for the tilt series. |
Tilt Axis |
TiltSeries.tilt_axis |
Rotation angle in degrees. |
Tilt Range |
TiltSeries.tilt_range |
Total tilt range in degrees. |
Tile Step |
TiltSeries.tiltstep |
Tilt step in degrees. |
Tilting Scheme |
TiltSeries.tilting_scheme |
The order of stage tilting during acquisition of the data. |
Total Flux |
TiltSeries.total_flux |
Number of electrons reaching the specimen in a square Angstrom area for the entire tilt series. |
Binning from Frames |
TiltSeries.binning_from_frames |
Describes the binning factor from frames to tilt series file. |
Series is Aligned |
No API field |
True or false, indicating whether the tilt series images have been transformed to account for the tomographic alignment. |
Related EMPIAR Entry |
TiltSeries.related_empiar_entry |
EMPIAR dataset identifier If a tilt series is deposited into EMPIAR. |
Tomogram
Portal Metadata |
API Expression |
Definition |
|---|---|---|
Reconstruction Software |
Tomogram.reconstruction_software |
Name of software used for reconstruction. |
Reconstruction Method |
Tomogram.reconstruction_method |
Reconstruction method, e.g. Weighted back-projection, SART, SIRT. |
Processing Software |
Tomogram.processing_software |
Processing software used to derive the tomogram. |
Available Processing |
Tomogram.processing |
Description of additional processing used to derive the tomogram, e.g. denoised. |
Smallest Available Voxel Spacing |
|
Smallest voxel spacing of the available tomograms. |
Size (x, y, z) |
|
Comma separated x,y,z dimensions of the unscaled tomogram. |
Fiducial Alignment Status |
Tomogram.fiducial_alignment_status |
Fiducial Alignment status: True = aligned with fiducial, False = aligned without fiducial. |
Ctf Corrected |
Tomogram.ctf_corrected |
Whether this tomogram is contrast transfer function corrected. |
Affine Transformation Matrix |
Tomogram.affine_transformation_matrix |
The flip or rotation transformation. |
Run Overview Page¶
The summary on the Run Overview page shows a subset of the run metadata as well as the number and type of files in the run and the tilt series quality score. The View Tomogram button opens the associated tomogram along with annotations using an instance of Neuroglancer in the browser. The annotations displayed with the tomogram are manually chosen to display as many annotations as possible without overlap or occlusion.
The table on a Run Overview page contains an overview of the annotations for the tomograms in this run. Each annotation has an Annotation ID, which is assigned by the Portal and is subject to change in the rare case where the annotation data needs to be re-ingested in the Portal. In addition to the Annotation ID, the table shows the object annotated, the type of the annotation, such as segmentation or point, the method used for annotation, such as manual or automated, and metrics for assessing the annotation.
Run Download Options¶
The Download ... button on the Run Overview page opens a dialog for downloading tomograms or instructions for downloading all annotations or all run data. Individual tomograms can be downloaded directly in the browser as MRC or OME-Zarr files following selection of which tomogram to download based on the tomogram sampling and processing. The All Annotations option provides instructions on using Amazon Web Services Command Line Interface or the Portal API to download.
You can also use the Portal API to download all run data as a folder using the Run ID in the download dialog or in the header on the Run Overview page. For example, to download all run data for Run ID 645 into your current working directory, use the below code snippet:
from cryoet_data_portal import Client, Run
client = Client()
run = Run.get_by_id(client, 645)
run.download_everything()
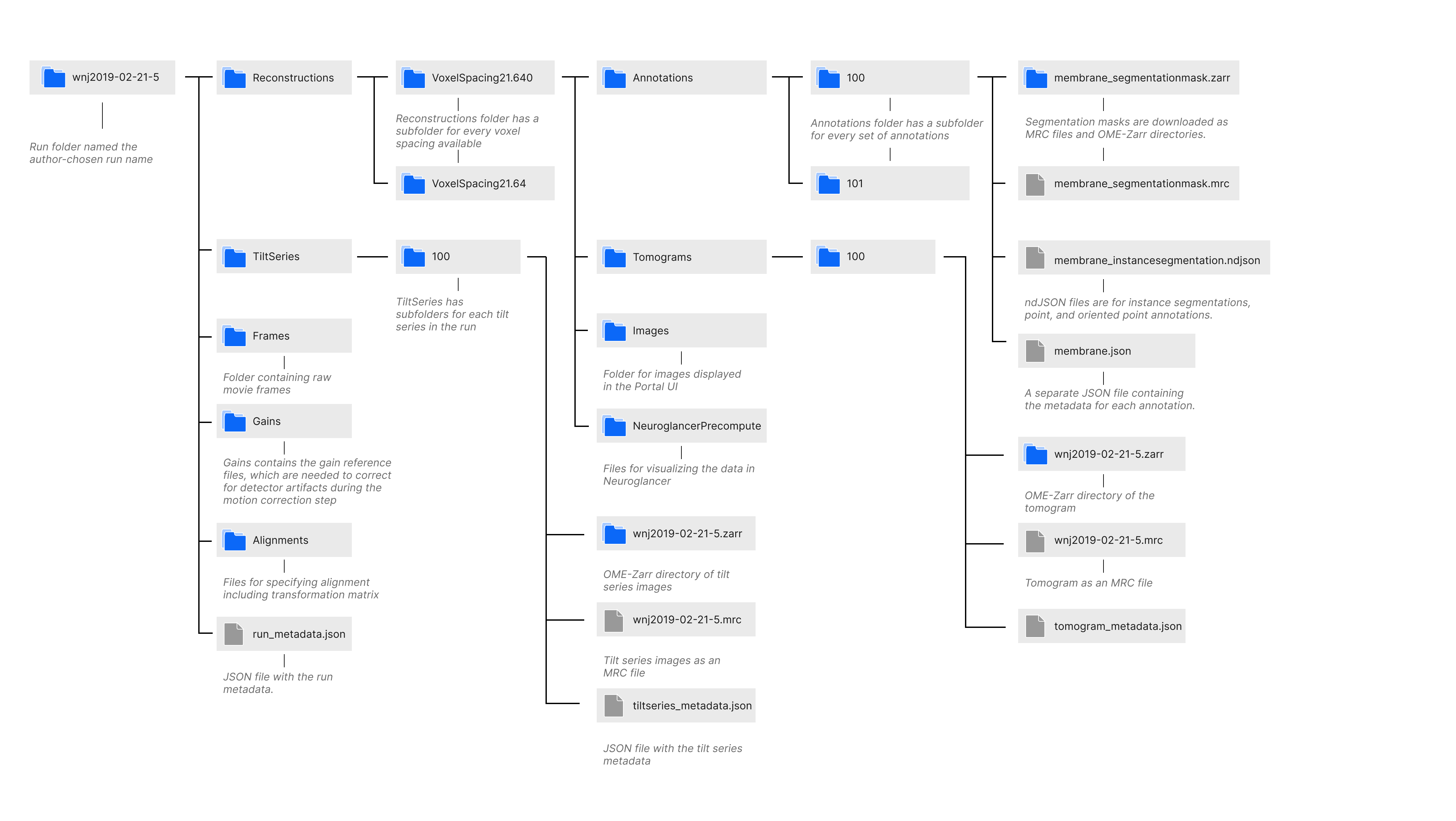
Runs are downloaded as folders named the author-chosen run name. As shown in the diagram above, the folder contains subfolders named Alignments, Frames, Gains, Reconstructions, and TiltSeries; and a JSON file named run_metadata.json containing the run metadata. The Alignments folder contains a JSON file for every alignment that specifies the alignment parameters including the affine transformation matrix. The Frames and Gains folders contain the raw movie frames and the gain files for correcting for the detector’s sensitivity, respectively. The TiltSeries folder contains the tilt series images as an MRC file and OME-Zarr directory as well as a JSON file with the tilt series metadata. The Reconstructions folder has a subfolder for every voxel spacing available, and each of those folders contains subfolders NeuorglancerPrecompute, which has files for visualizing the data in Neuroglancer; Images, which contains the key photos of the run displayed on the Portal; Tomograms, which has the tomograms associated with the run as an MRC file and OME-Zarr directory along with tomogram metadata and metadata for visualizing the tomogram with Neuroglancer; and Annotations. More details on the Annotation folder file structure is found in the documentation below.
Annotations¶
Annotations are summarized in a table on Run Overview pages. Each annotation has an Annotation ID, which is assigned by the Portal and is subject to change in the rare case where the annotation data needs to be re-ingested in the Portal. Annotation IDs start with AN (e.g. AN-2480; note that only the numeric part is supported in the API). Each annotation labels exactly one type of object, such as ribosome or membrane, indicated by the Object Type column of the table. For every object, there is one type of annotation per entry in the table indicated in the Object Shape Type column. The options are Segmentation for semantic segmentation masks, Instance Segmentation for segmentation masks where each individual object is labeled with its own color, Point for point annotations, and Oriented Point for point annotations that have an associated rotation matrix. The method used for generating the annotation is displayed for each annotation with manual meaning the annotations were created by hand, automated meaning automated tools or algorithms without supervision were used, and hybrid meaning the annotations were generated using a combination of automated and manual methods.
Annotations also have an optional precision field, which is the percentage of true positives among the total number of positive predictions where a value of 100% means everything found is actually the object of interest, and a recall field, which is the percentage of true positives among the actual number of objects where a value of 100% meaning all objects of interest were found. The Precision and Recall fields can only be calculated by comparing with a ground truth annotation, so for many annotations on the Portal, this field is marked NA for not available.
Authors may also utilize the Ground Truth flag on entries in the annotation table. The Ground Truth flag indicates that this annotation is endorsed by the author for use as training or validation data for machine learning models.
Each annotation has its own metadata, which can be viewed using the info icon on the entry in the annotations table. These metadata are defined in the tables below including their mapping to attributes in the Portal API:
Annotation Overview
Portal Metadata |
API Expression |
Definition |
|---|---|---|
Annotation ID |
Annotation.id |
Numeric identifier assigned by the Portal. |
Annotation Authors |
Annotation.authors |
Authors of this annotation. |
Publication |
Annotation.annotation_publication |
DOIs for publications that describe the dataset. |
Deposition Date |
Annotation.deposition_date |
Date when an annotation set is initially received by the Data Portal. |
Release Date |
Annotation.release_date |
Date when annotation data is made public by the Data Portal. |
Last Modified Date |
Annotation.last_modified_date |
Date when an annotation was last modified in the Data Portal. |
Method Type |
Annotation.method_type |
The method type for generating the annotation (e.g., manual, hybrid, automated). |
Annotation Method |
Annotation.annotation_method |
Describes how the annotation is made, e.g., Manual, crYoLO, Positive Unlabeled Learning, template matching. |
Annotation Software |
Annotation.annotation_software |
Software used for generating this annotation. |
Annotation Object
Portal Metadata |
API Expression |
Definition |
|---|---|---|
Object Name |
Annotation.object_name |
Name of the object being annotated, e.g., ribosome, nuclear pore complex, actin filament, membrane. |
Object ID |
Annotation.object_id |
Gene Ontology Cellular Component identifier or UniProtKB accession for the annotation object. |
Object Count |
Annotation.object_count |
Number of objects identified. |
Object Shape Type |
AnnotationFile.shape_type |
Description of whether this is a Point, OrientedPoint, or SegmentationMask file. |
Object State |
Annotation.object_state |
Additional information about the annotated object not captured by the gene ontology (e.g., open or closed state for molecules). |
Object Description |
Annotation.object_description |
Description of the annotated object, including additional information not covered by the Annotation object name and state. |
Annotation Confidence
Portal Metadata |
API Expression |
Definition |
|---|---|---|
Ground Truth Status |
Annotation.ground_truth_status |
Whether an annotation is considered ground truth, as determined by the annotation author. |
Ground Truth Used |
Annotation.ground_truth_used |
Annotation filename used as ground truth for precision and recall. |
Precision |
Annotation.confidence_precision |
Percentage of annotation objects being true positive. |
Recall |
Annotation.confidence_recall |
Percentage of true positives being annotated correctly. |
Visualizing Annotations with Tomograms in Neuroglancer¶
There is no definitive rule for which annotations are displayed with a tomogram in Neuroglancer by default. The annotations are manually chosen by the data curation team to display as many annotations as possible without overlap or occlusion. For example, when the cytoplasm is annotated as a whole, it would occlude other annotations, such as protein picks. When there is a ground truth and predicted annotation, the ground truth annotation is displayed by default. Authors contributing data can specify the desired default annotations during the submission process.
The CryoET Data Portal napari plugin can be used to visualize tomograms, annotations, and metadata. Refer to this documentation to learn about how to use the plugin and to this page to learn more about napari and CryoET Data Portal.
Annotation Download Options¶
Individual entries in the annotations table can be downloaded using the Download button on the right side of the table.
Instance Segmentations, Oriented Points, and Points all can be downloaded directly in the browser as Newline Delimited JSON (ndJSON) files, where each line in the file is its own JSON. The download dialog also has instructions for downloading using curl, Amazon Web Services Command Line Interface or the Portal API. In all cases, the JSON entries have a type field with instancePoint, orientedPoint, and point for Instance Segmentations, Oriented Points, and Points, respectively, and a location field with the x, y, z coordinates. For Instance Segmentations, there is also an instance_id to group points into geometric segmentation masks. For Oriented Points, there is also an xyz_rotation_matrix field with a 3x3 rotation matrix corresponding to each point.
Semantic segmentation masks can downloaded using Amazon Web Services Command Line Interface or the Portal API as MRC files or OME-Zarr directories. When downloading all annotations on a Run Overview page, both the MRC file and the OME-Zarr directory will be downloaded for each segmentation mask.