First stable iteration of Census (SOMA) PyTorch loaders
NOTICE (2025): Since this article’s publication, the Census PyTorch loaders were superseded by TileDB-SOMA-ML and removed from the cellxgene_census.experimental API. Our revised Training a PyTorch Model notebook reflects the new approach, while the outdated information below remains for historical interest.
Published: July 11th, 2024
Updated: July 19th, 2024. Figure 3 has been improved for readability.
By: Emanuele Bezzi, Pablo Garcia-Nieto, Prathap Sridharan, Ryan Williams
The Census team is excited to share the release of Census PyTorch loaders that work out-of-the-box for memory-efficient training across any slice of the >70M cells in Census.
In 2023, we released a beta version of the loaders and we have observed interest from users to utilize them with Census or their own data. For example Wolf et al. performed comparisons across different training approaches and found our loaders to be ideal for uncached training of Census data, albeit with some caveats.
We have continued the development of the loaders in collaboration with our partners at TileDB, and we are happy to announce this release as the first stable iteration. We hope the loaders can accelerate the development of large-scale models of single-cell data by leveraging the following main features:
Out-of-the-box training on all or any slice of Census data.
Efficient memory usage with out-of-core training.
Calibrated shuffling of observations (cells).
Cloud-based or local data access.
Increased training speed.
Custom data encoders.
Keep on reading for usage and more details on the main loader features.
Census PyTorch loaders usage
The loaders are ready to use for PyTorch modeling via the specialized Data Pipe ExperimentDataPipe, which takes advantage of the out-of-core data access TileDB-SOMA offers.
Please follow the Training a PyTorch Model tutorial for a full reproducible example to train a logistic regression on cell type labels.
In short, the following shows you how to initialize the loader to train a model on a small subset of cells. First, you can initialize a ExperimentDataPipe to train a model on tongue cells as follows:
import cellxgene_census.experimental.ml as census_ml
import cellxgene_census
import tiledbsoma as soma
experiment = census["census_data"]["homo_sapiens"]
experiment_datapipe = census_ml.ExperimentDataPipe(
experiment,
measurement_name="RNA",
X_name="raw",
obs_query=soma.AxisQuery(value_filter="tissue_general == 'tongue' and is_primary_data == True"),
obs_column_names=["cell_type"],
batch_size=128,
shuffle=True,
)
Then you can perform any PyTorch operations and training.
# Splitting training and test sets
train_datapipe, test_datapipe = experiment_datapipe.random_split(weights={"train": 0.8, "test": 0.2}, seed=1)
# Creating data loader
experiment_dataloader = census_ml.experiment_dataloader(train_datapipe)
# Training a PyTorch model
device = torch.device("cuda") if torch.cuda.is_available() else torch.device("cpu")
model = MODEL().to(device)
model.train()
Census PyTorch loaders main features
Out-of-the-box training on all or any slice of Census data
Since the ExperimentDataPipe inherits from the PyTorch Iterable-style DataPipe it can be readily used with PyTorch models.
The single-cell expression data is encoded in numerical tensors, and for supervised training the cell metadata can be automatically transformed with a default encoder, or with custom user-defined encoders (see below).
Efficient memory usage with out-of-core training
Thanks to the underlying backend of Census — TileDB-SOMA — the PyTorch loaders take advantage of incremental data materialization of fixed and small size to keep memory usage constant throughout training.
In addition, data is eagerly fetched while batches go through training so that compute is never idle or waiting for data to be loaded. This feature is particularly useful when fetching Census data directly from the cloud.
Memory usage is defined by the parameters soma_chunk_size and shuffle_chunk_count - see below for a full description on how these should be tuned.
Calibrated shuffling of observations (cells)
Shuffling along efficient out-of-core data fetching is a challenge. In general, increasing randomness of shuffling leads to slower data fetching.
In the first iteration of the loaders, shuffling was done through large blocks of data of user-defined size. This shuffling strategy led to non-random distribution of observations per training batch, becasue Census has a non-random data structure (observations from the same datasets are adjacent to one another) thus training loss was unstable (Figure 1).
Now we have implemented a scatter-gather approach, whereby multiple chunks of data are fetched randomly from Census, then a number of chunks are concatenated into a block and all observations within the block are randomly shuffled. Adjusting the size and number of chunks per block leads to well-calibrated shuffling with stable training loss (Figure 2) while maintaining efficient data fetching (Figure 3).
The balance between memory usage, efficiency, and level of randomness can be adjusted with the parameters soma_chunk_size and shuffle_chunk_count. Increasing shuffle_chunk_count will improve randomness, as more scattered chunks will be collected before the pool is randomized. Increasing soma_chunk_size will improve I/O efficiency while decreasing it will improve memory usage. We recommend a default of soma_chunk_size=64, shuffle_chunk_count=2000 as we determined this configuration yields a good balance.
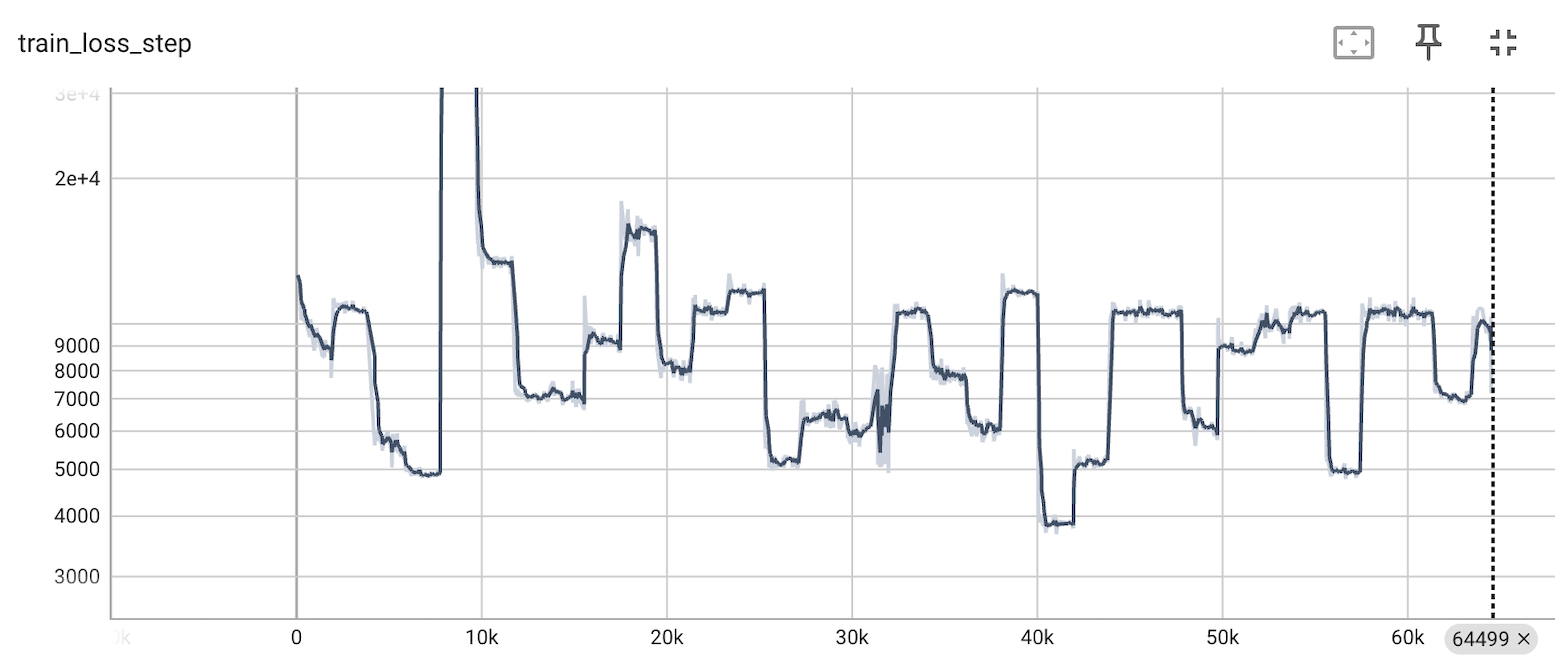
Figure 1. Training loss was unstable with the previous shuffling strategy. Based on a trial scVI run on 64K Census cells.
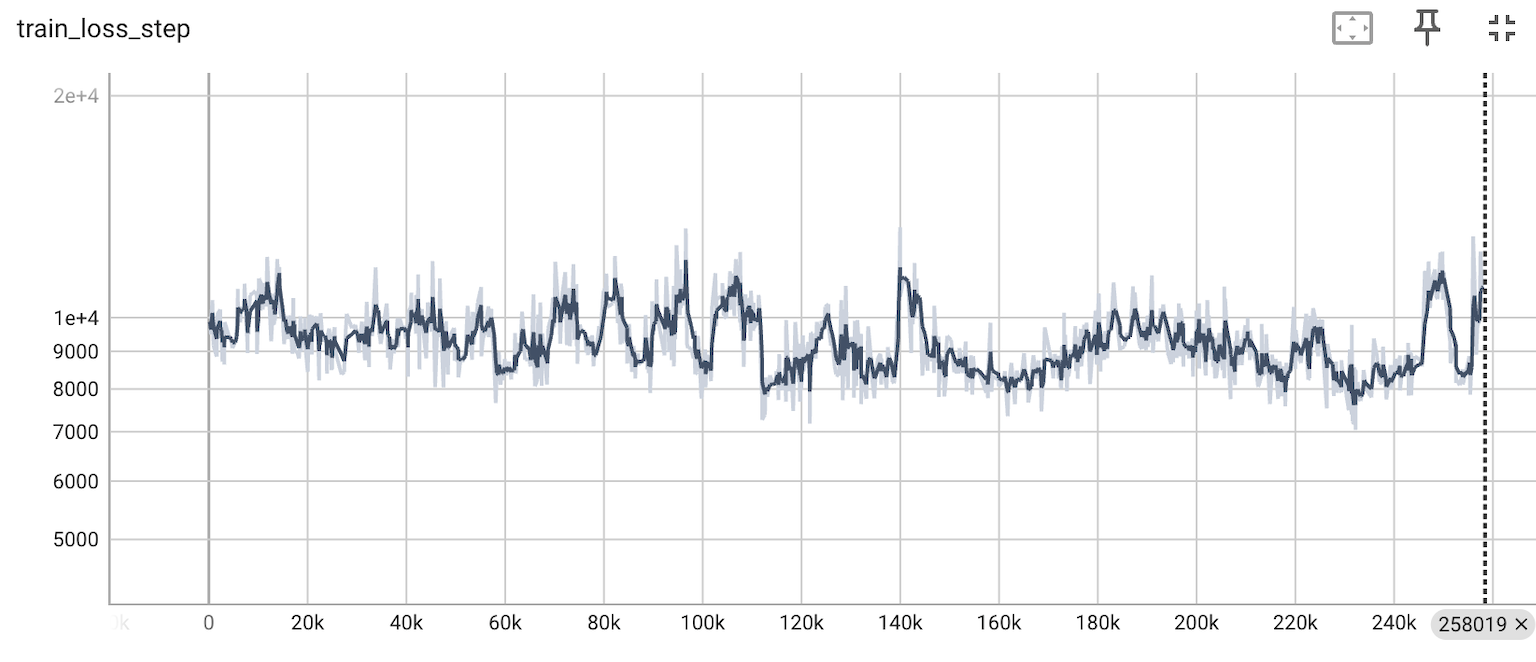
Figure 2. Training loss is well-calibrated with the current scatter-gather shuffling strategy. Based on a trial scVI run on 250K Census cells.
Increased training speed
We have made improvements to the loaders to reduce the amount of data transformations required from data fetching to model training. One such important change is to encode the expression data as a dense matrix immediately after the data is retrieved from disk/cloud.
In our benchmarks, we found that densifying data increases training speed while maintaining relatively constant memory usage (Figure 3). For this reason, we have disabled the intermediate data processing in sparse format unless Torch Sparse Tensors are requested via the ExperimentDataPipe parameter return_sparse_X.
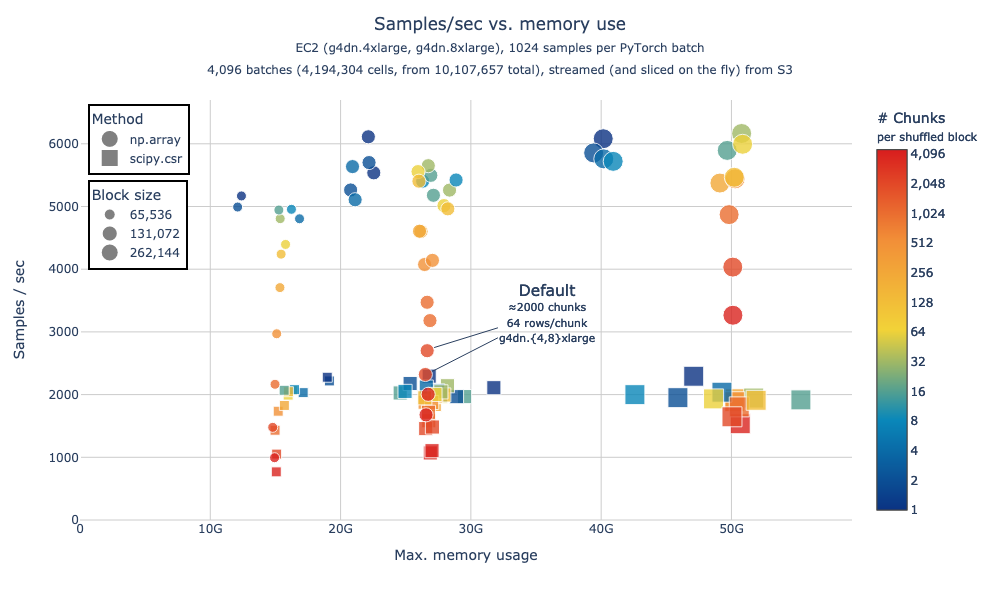
Figure 3. Benchmark of memory usage and speed of data processing during modeling, default parameters lead to ≈2,500 samples/sec with 27GB of memory use. The benchmark was done processing 4M cells out of a 10M-cell Census, with data streamed from the cloud (S3). “Method” indicates the expression matrix encoding: circles are dense (“np.array”, now the default behavior) and squares are sparse (“scipy.csr”). Size indicates the total number of cells per processing block (max cells materialized at any given time) and color is the number of individual randomly grabbed chunks composing a processing block; higher chunks per block lead to better shuffling. Data was fetched until modeling step, but no model was trained.
We repeated the benchmark in Figure 3 in different conditions encompassing varying number of total cells and multiple epochs, please follow this link for the full benchmark report and code..
When comparing dense vs sparse processing in an end-to-end training exercise with scVI, we also observed slight increased speed with the dense approach and comparable memory usage to sparse processing (Figure 4). However in this full training example the differences were less substantial, highlighting that other model-specific factors during the training phase will contribute to memory and speed performance.
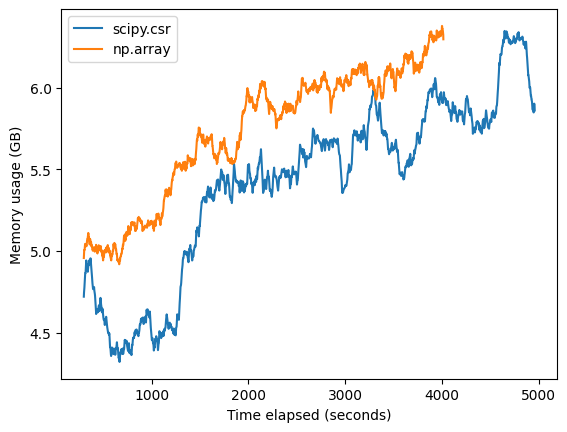
Figure 4. Trial scVI training run with default parameters of the Census Pytorch loaders, highlighting increased speed of dense vs sparse data processing. Training was done on 5684805 mouse cells for 1 epoch on a g4dn.16xlarge EC2 machine.
Custom data encoders
For maximum flexibility, users can provide custom encoders for the cell metadata enabling custom transformations or interactions between different metadata variables.
To use custom encoders you need to instantiate the desired encoder via the Encoder class and pass it to the encoders parameter of the ExperimentDataPipe.
